
The Certificate of Analysis (COA) is a signed document that includes the storage temperature, expiration date and quality controls for an individual lot. Reaction mixture included 1 g of DNA and 1 L of each restriction enzyme to a total volume of 30 L for triple digestion, as per recommended protocol. Locate commercially available restriction enzymes by category, name, recognition sequence, or overhang. For single restriction enzyme digestions, reaction mixture included 1 g of DNA and 1 l of restriction enzyme to a total volume of 20 L. This product is related to the following categories: Restriction Endonucleases: N-O, Time-Saver Qualified Restriction Enzymes Products This product can be used in the following applications: Restriction Enzyme Digestion using Anza 1 NotI, Anza 16 HindIII and Anza 15 XmaJI.
coli strain that carries the cloned NotI gene from Nocardia otitidis-caviarum (ATCC 14630). Engineered with performance in mind, HF restriction enzymes are fully active under a broader range of conditions, minimizing off-target products, while offering flexibility in experimental design.įor details on NEB’s quality controls for restriction endonucleases, visit our Restriction Enzyme Quality page.Īn E. Note that NotI is a methylation-sensitive enzyme, i.e.
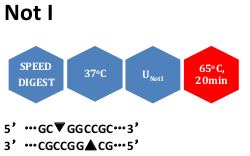
HF enzymes are all Time-Saver qualified and can therefore cut substrate DNA in 5-15 minutes with the flexibility to digest overnight without degradation to DNA. NEB began switching our BSA-containing reaction buffers in April 2021 to buffers containing Recombinant Albumin (rAlbumin) for restriction enzymes and some DNA. The nicking enzymes cut ('nick') only one strand in a DNA duplex. HF enzymes also exhibit dramatically reduced star activity. Not I cleaves prokaryotic genomic DNA to generate. High Fidelity (HF) Restriction Enzymes have 100% activity in rCutSmart Buffer single-buffer simplicity means more straightforward and streamlined sample processing. Plasmid DNA (6,215 bp) and purified PCR product (1.6 kb) were digested using Anza 11 EcoRI, Anza 12 XbaI, and Anza 1 NotI. It is one of the two known enzymes recognizing an octameric sequence comprised solely of G and C residues. Activity data in Promega buffers are not available for all enzymes.NotI has a High Fidelity version NotI-HF ® ( NEB #R3189). Once you have selected an enzyme, you can select a second enzyme and retrieve information on suitable buffers for double digests. Search by specific 3' or 5' overhang, or list all blunt enzymes. The frequency of cutting in a random DNA sequence for a given restriction enzyme is once per every 4n, where n is the number of bases in the restriction. In addition, the universal Tango buffer is provided for convenience in double digestions. NotI has a High Fidelity version NotI-HF (NEB R3189). Ad2 DNA digested with NotI, 0.7 agarose, 7 cleavage sites conventional restriction endonucleases are a large collection of high quality restriction enzymes, optimized to work in one of the buffers of the Five Buffer System. W = A or T (weak interactions: 2 H bonds) Reduce Star Activity with High-Fidelity Restriction Enzymes. S = G or C (strong interactions: 3 H bonds) The following IUPAC codes can be used when the wildcard match is turned ON: Engineered with performance in mind, HF restriction enzymes are fully active under a broader range of conditions, minimizing off-target products, while offering. 64.90 - 244.50 Products 3 Description Description Ad2 DNA digested with NotI, 0. Only exact matches will be returned when the "wildcard match" is turned OFF. Thermo Scientific NotI (10 U/L) The NotI restriction enzyme recognizes GCGGCCGC sites and cuts best at 37C in O buffer (Isoschizomers: CciNI). Select "Sequence", and enter the recognition sequence into the search box. NotI Digestions Prepare a stock reaction mix that is 2X in restriction enzyme reaction buffer including bovine serum albumin (BSA) (reaction buffer) and. High Fidelity (HF) Restriction Enzymes have 100 activity in CutSmart Buffer single-buffer simplicity means more straightforward and streamlined sample.
Select "Name", and enter the name of your enzyme into the search box. You can find an enzyme by entering the name, recognition sequence or overhang sequence.


 0 kommentar(er)
0 kommentar(er)
